Workflow for diffusion MRI
Table of contents
Automated processing pipeline for diffusion MRI
Processing baby diffusion MRI (dMRI) data is challenging due to the low and spatially-varying diffusion anisotropy, rendering standard analysis techniques developed for adult data inapplicable [1,2]. Here, we present a fully-automated processing pipeline for baby dMRI, tailored particularly to the data collected in the Baby Connectome Project (BCP).
The human brain is arguably the most complex system in biology and yet its macroscopic layout is nearly complete by the time of term birth. The infant brain develops rapidly during the first years of life, posing significant challenges to precise quantification of rapid dynamic changes that occur during this critical period of brain development.
The increasing availability of longitudinal baby MRI data, such as those acquired through the Baby Connectome Project (BCP) [3], affords unprecedented opportunity for precise charting of early brain developmental trajectories in order to understand normative and aberrant growth.
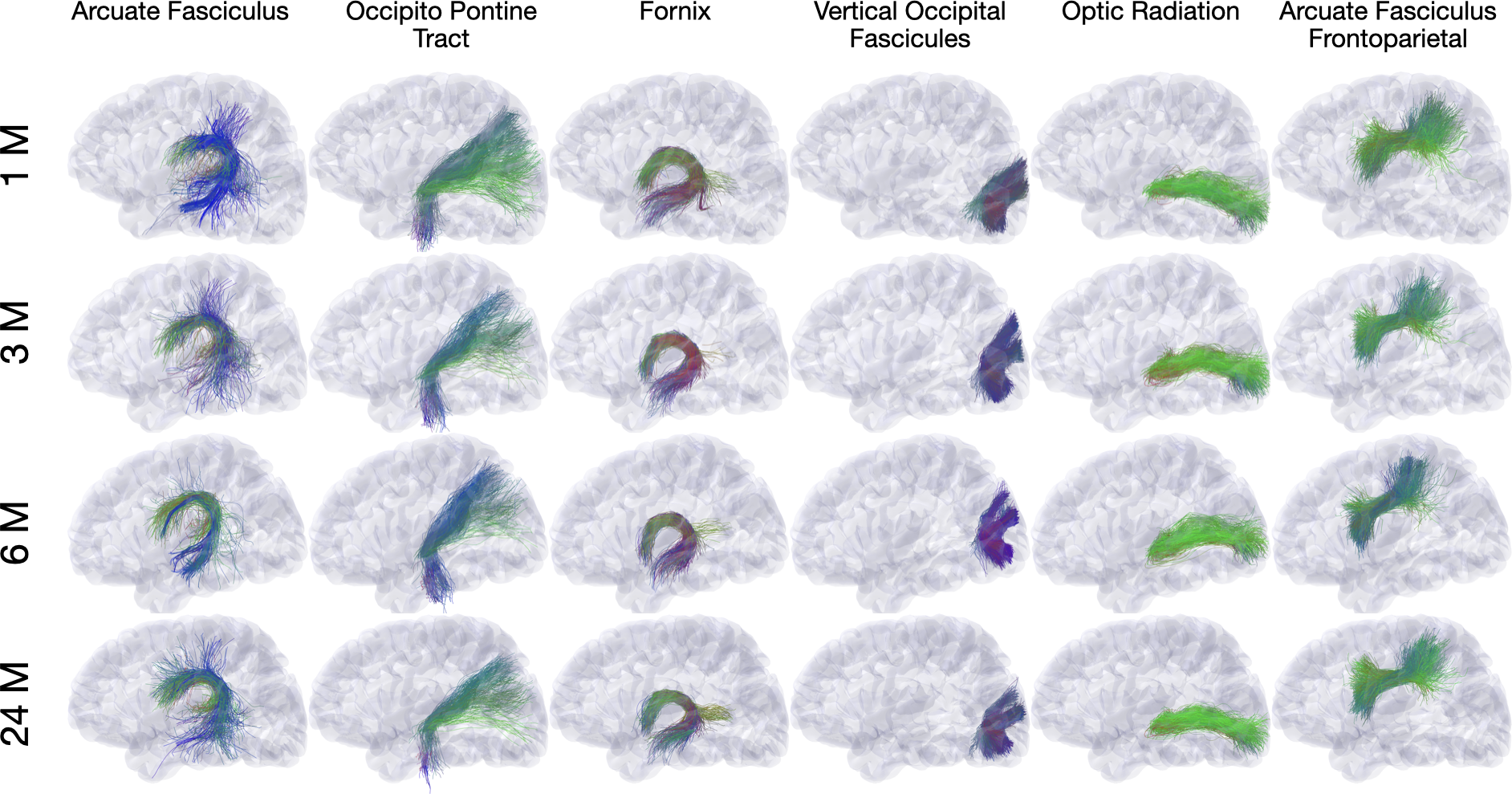
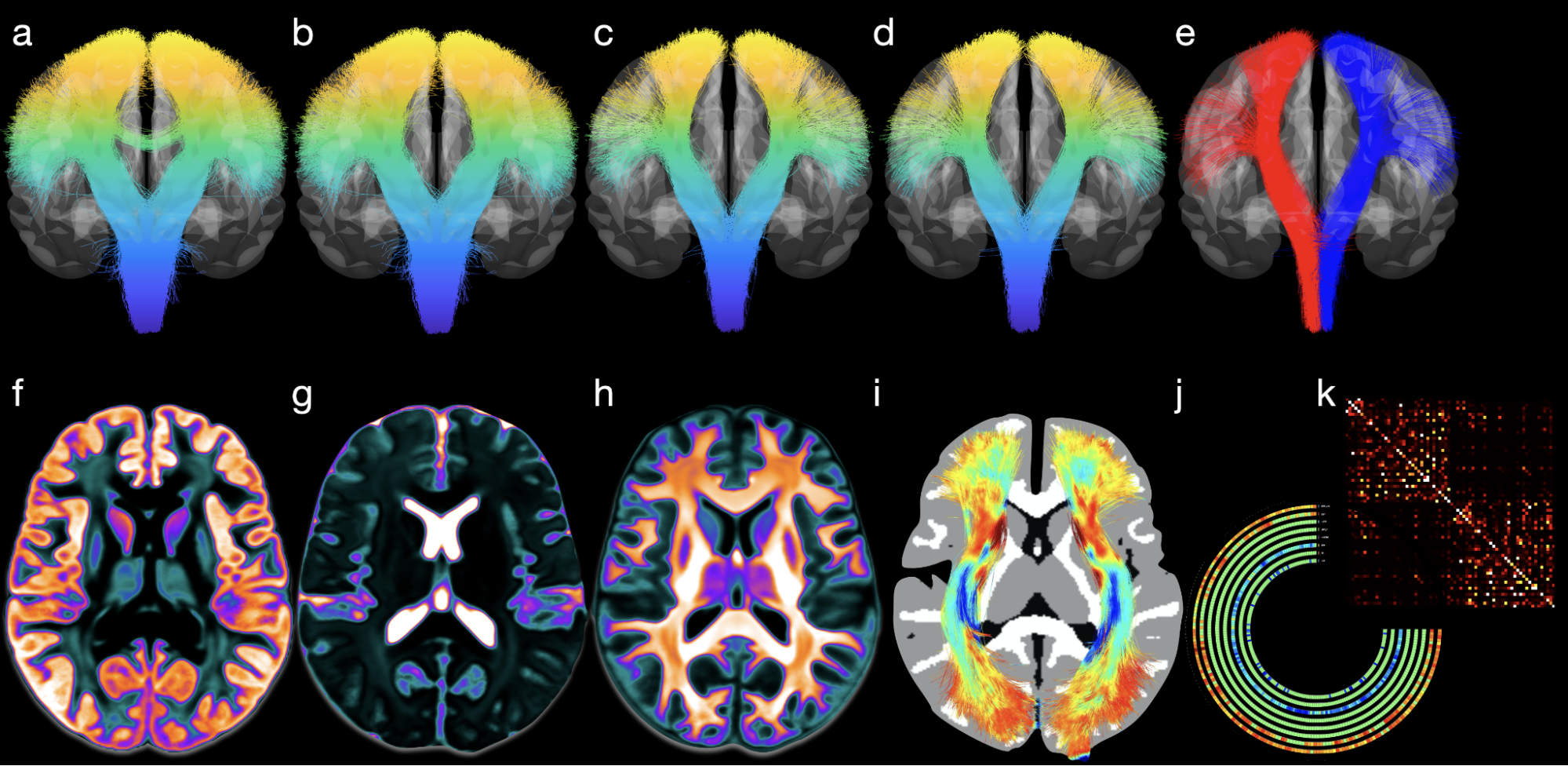
We describe here an automated processing pipeline for baby dMRI, consisting of (i) image quality control (IQC) for distinguishing between good and poor quality data, (ii) anatomy-guided correction for eddy-current and susceptibility-induced distortions, (iii) tissue microstructural analysis, and (iv) reconstruction of white matter (WM) pathways.
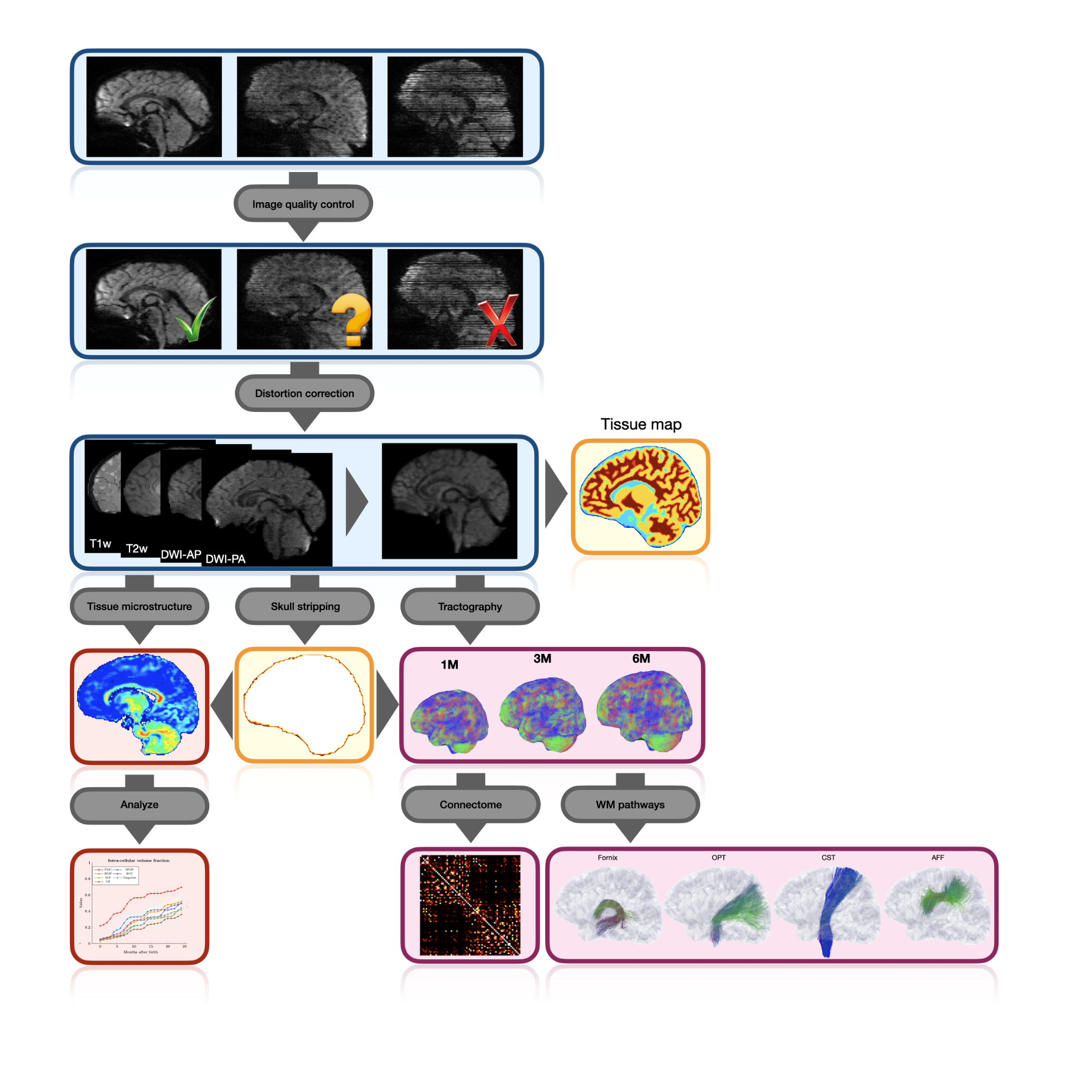
Wu, Y., Ahmad, S., Huynh, KM., Liu, S., Thung, K-H., Lin, W. & Yap, P. T. (2021). An Automated Processing Pipeline for Diffusion MRI in the Baby Connectome Project Proceedings of the International Society of Magnetic Resonance in Medicine (ISMRM) (Summa Cum Laude Award)
Computational medical imaging toolkit
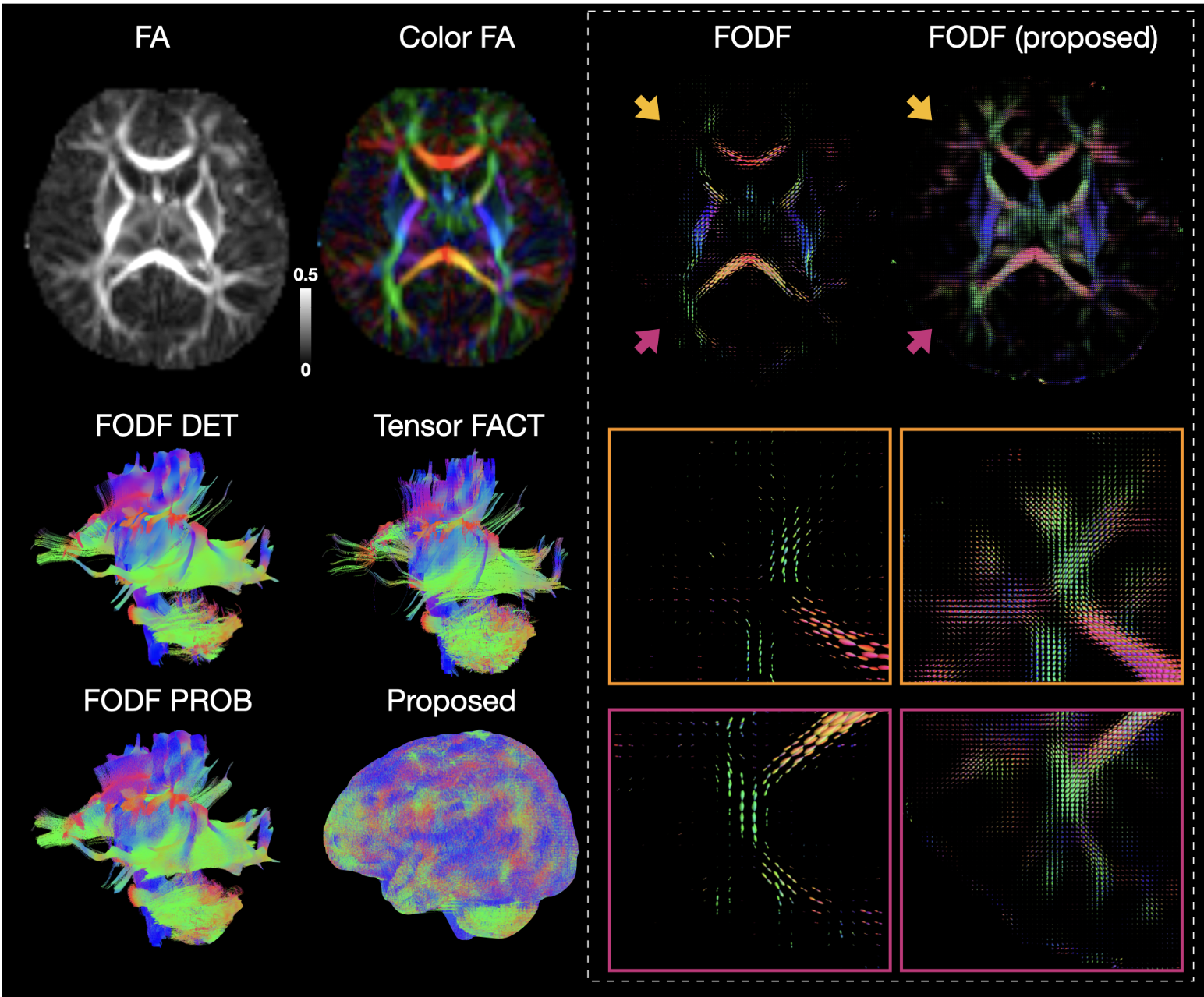
Processing and analyzing diffusion MRI (dMRI) data is important for uncovering the neural underpinnings of white matter (WM) development and degeneration across the human lifespan. Here, we introduce a robust and integrative toolkit, called Computational Medical Imaging (COMEDI), for processing, analyzing, and visualizing lifespan dMRI data.
Diffusion MRI (dMRI) is a primary tool for noninvasive and in vivo investigation of tissue microstructure and white matter (WM) pathways in the human brain1,2. Making sense of dMRI data often involves intensive processing, fine-grained analysis, and multi-perspective visualization. Despite the availability of many software packages for dMRI processing, few are effective for the entire human lifespan, particularly the first years of life where the diffusion characteristics are markedly different from adults. Here, we introduce a robust and integrative dMRI processing toolkit — Computational Medical Imaging (COMEDI) — that has been tested on dMRI data spanning the entire human lifespan from birth to 100 years of age. COMEDI covers recent advances in dMRI processing and analysis, including asymmetric fiber orientation distribution functions, cortico-cortical tractography with explicit consideration of gyral bias, advanced microstructural analysis, consistent bundle identification across the human lifespan, dedicated analysis pipelines for baby dMRI, and subject-specific whole-brain parcellation. COMEDI will be made publicly available as a non-commercial software package after the ISMRM meeting to support lifespan dMRI studies.
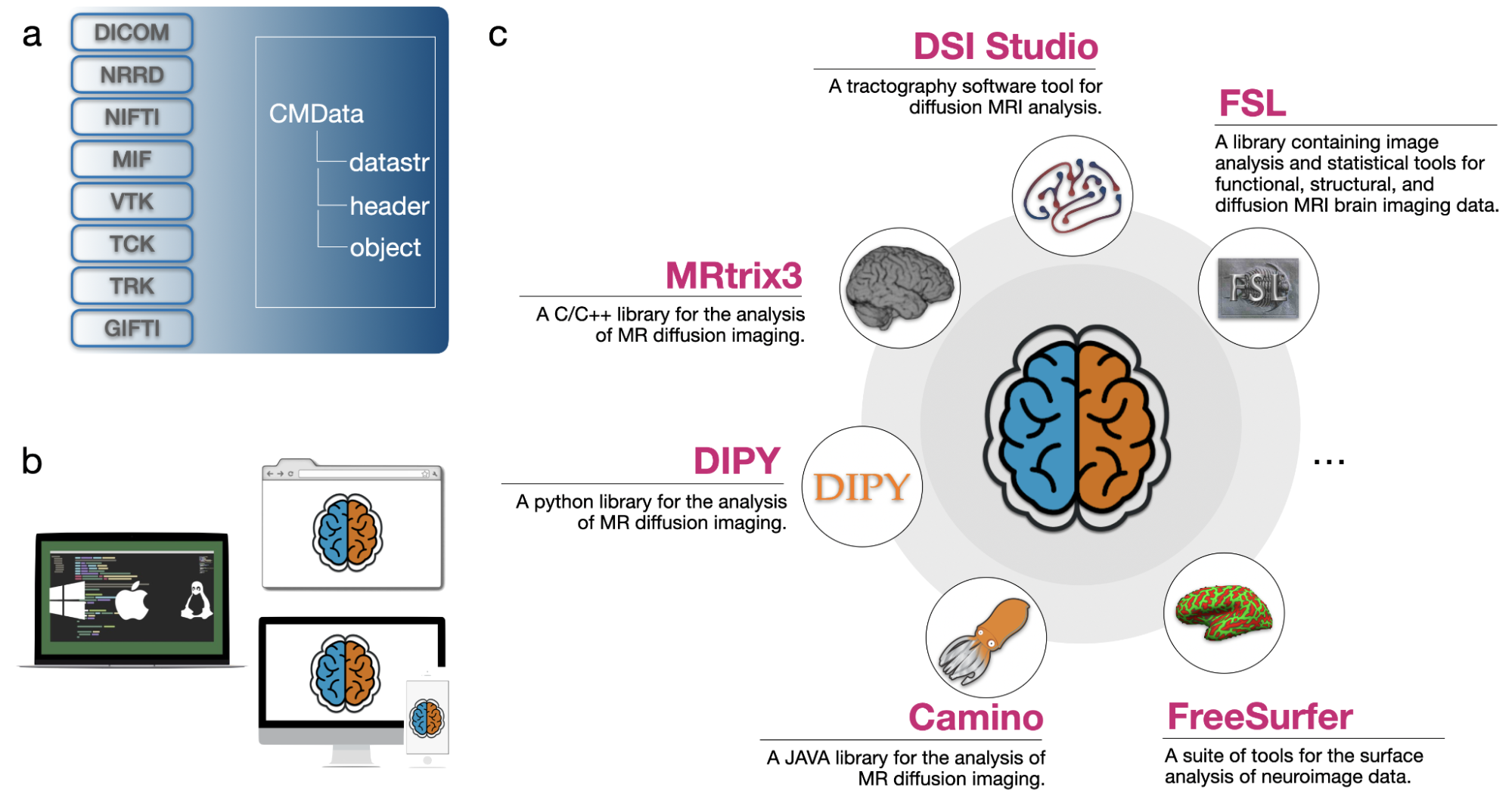
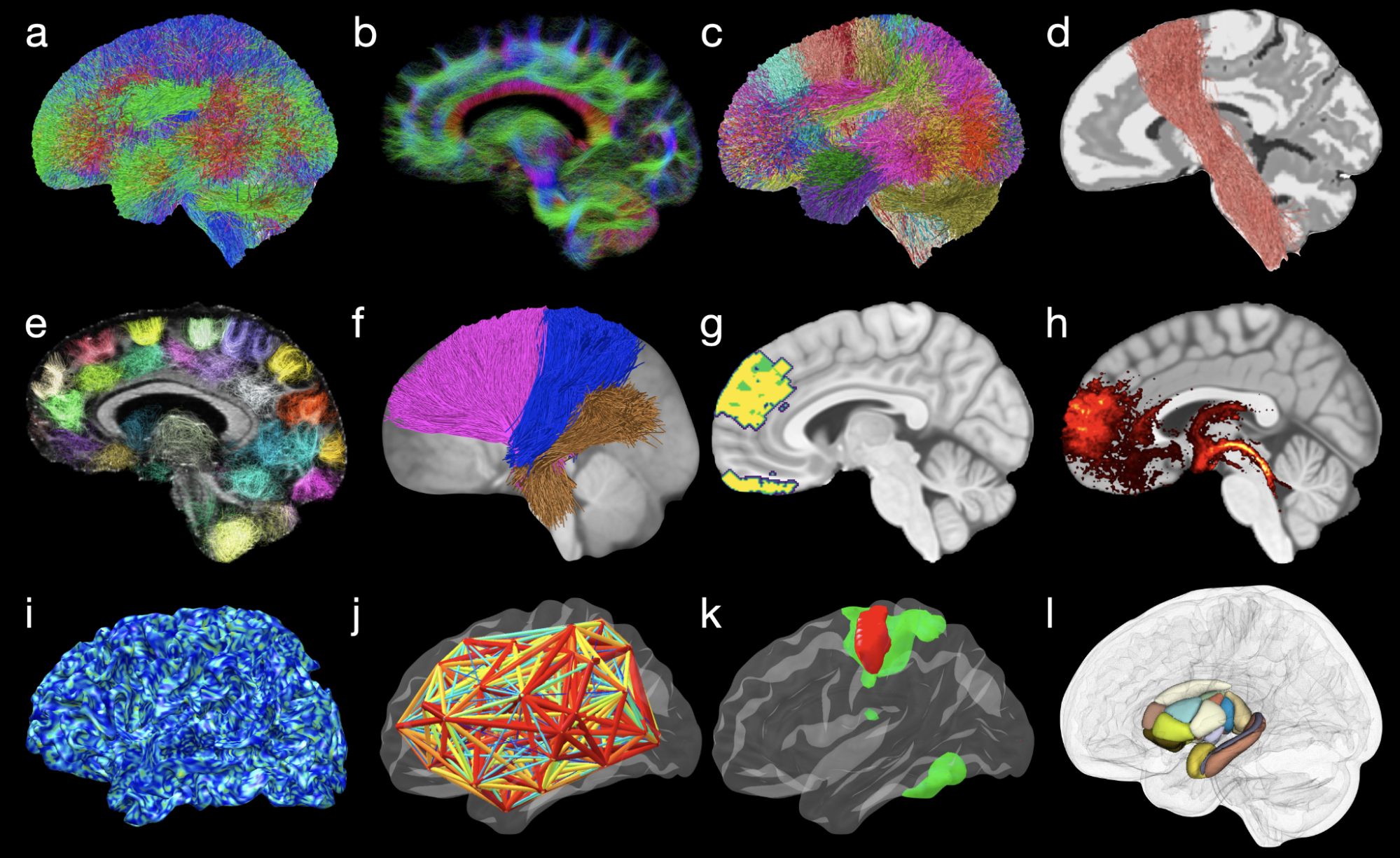
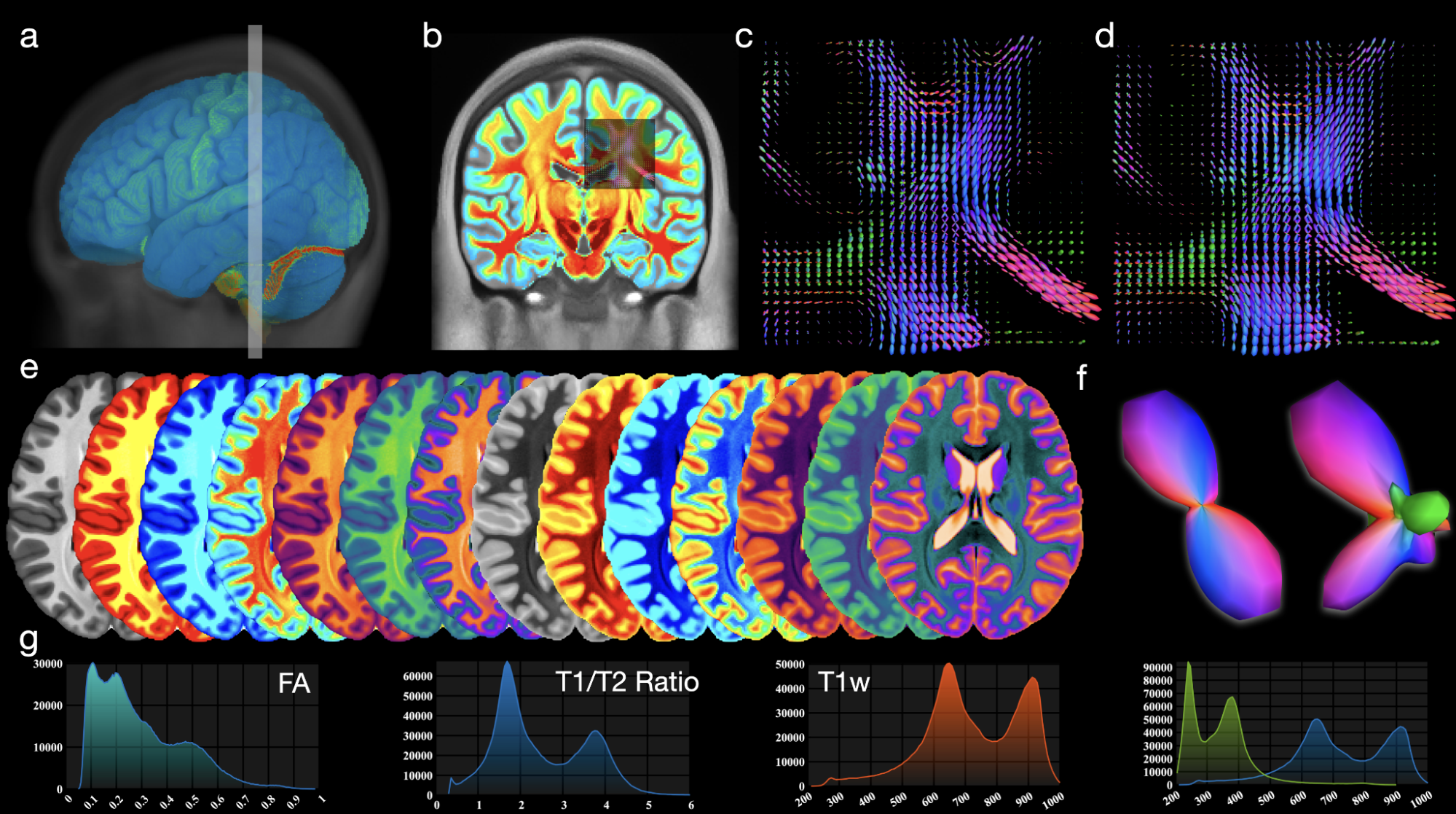
Wu, Y., Ahmad, S. Huynh, KM., Xu T. & Yap, P. T. (2022). COMEDI: A Toolkit for Lifespan Computational Diffusion MRI Proceedings of the International Society of Magnetic Resonance in Medicine (ISMRM)
Fast correction of eddy-current and susceptibility-induced distortions
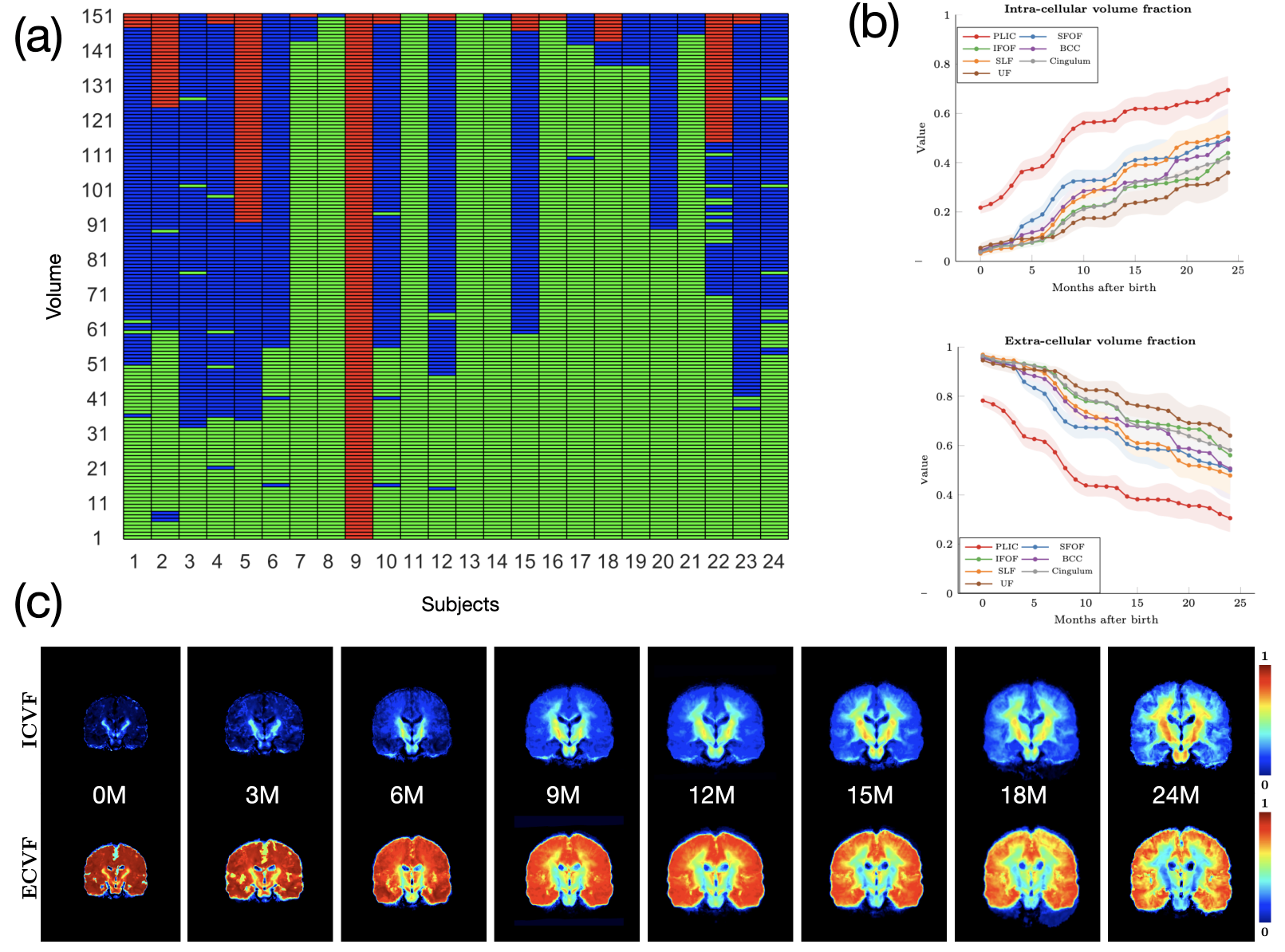
Diffusion MRI (dMRI) is typically time consuming as it involves acquiring a series of 3D volumes, each associated with a wave-vector in q-space that determines the diffusion direction and strength. The acquisition time is further increased when “blip-up blip-down” scans are acquired with opposite phase encoding directions (PEDs) to facilitate distortion correction. In this work, we show that geometric distortions can be corrected without acquiring with opposite PEDs for each wave-vector, and hence the acquisition time can be halved. Our method uses complimentary rotation-invariant contrasts across shells of different diffusion weightings. Distortion-free structural T1-/T2-weighted MRI is used as reference for nonlinear registration in correcting the distortions. Signal dropout and pileup are corrected with the help of spherical harmonics. To demonstrate that our method is robust to changes in image appearance, we show that distortion correction with good structural alignment can be achieved within minutes for dMRI data of infants between 1 to 24 months of age.
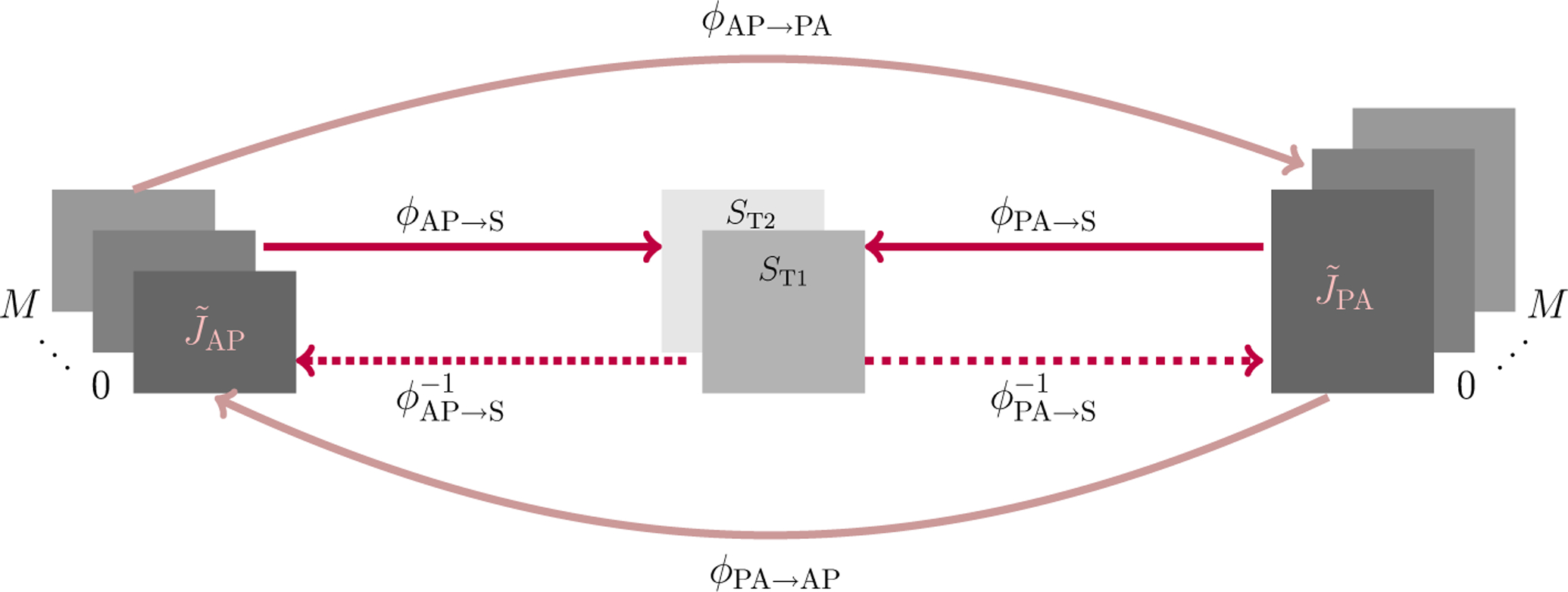
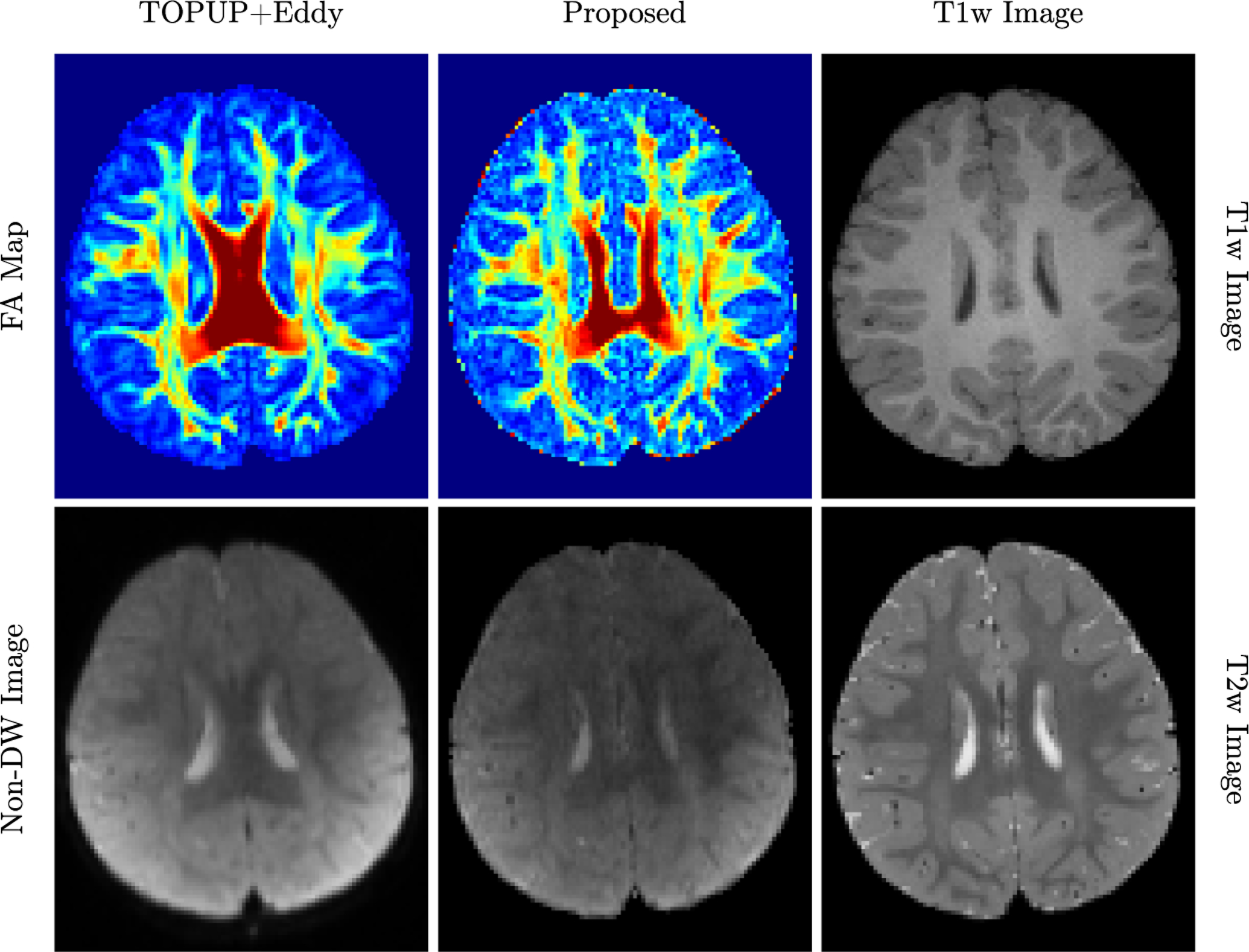
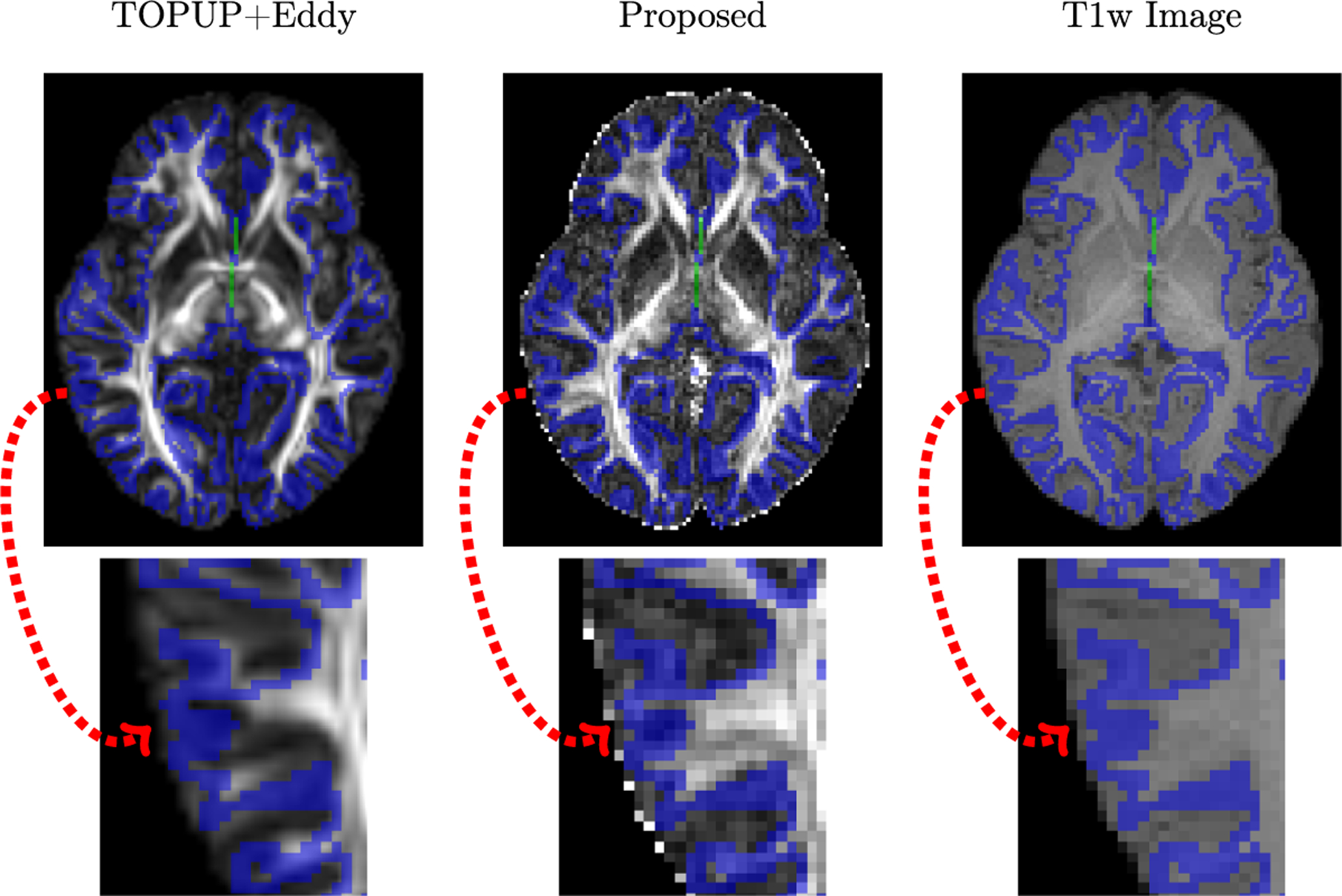
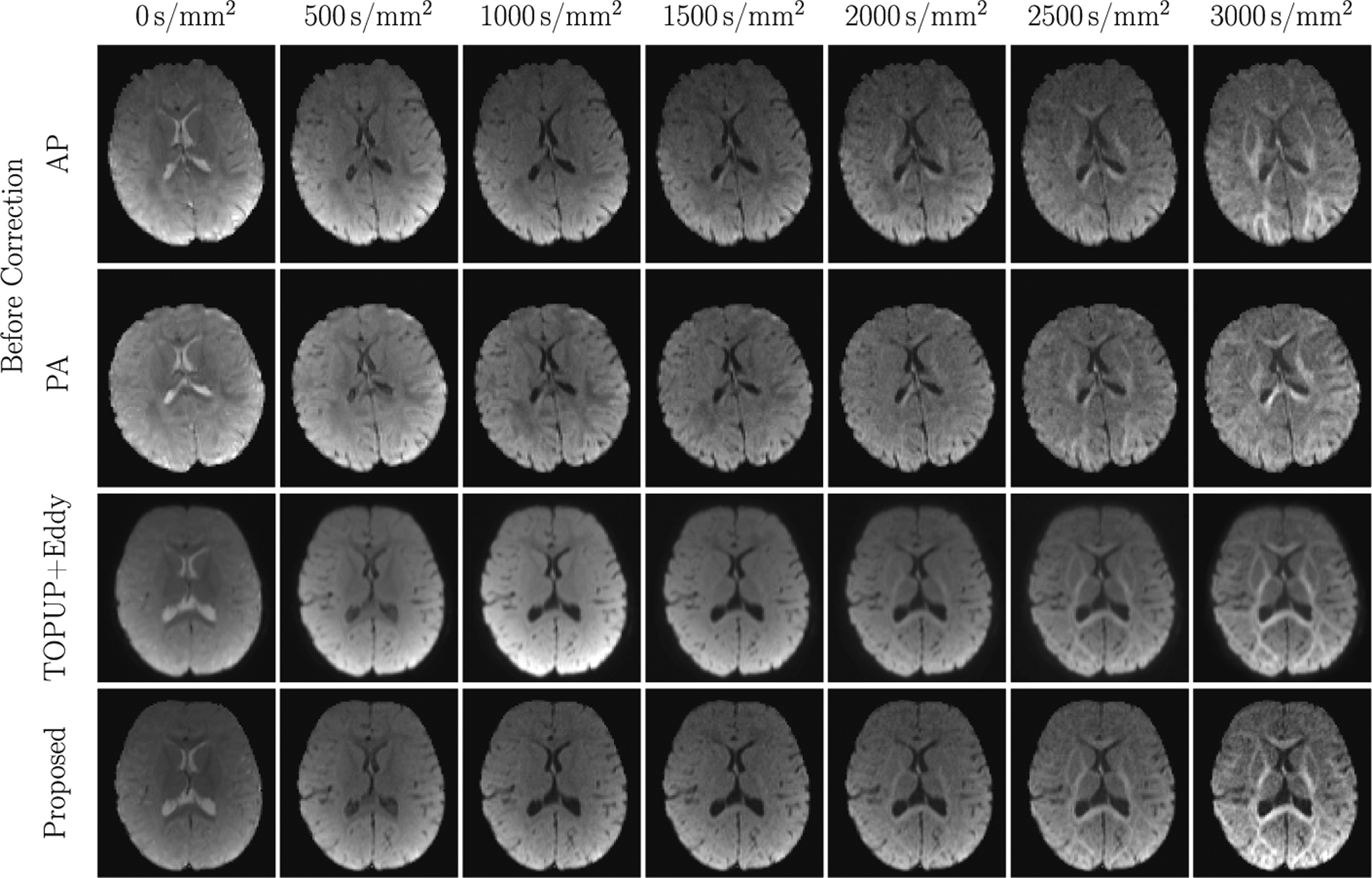
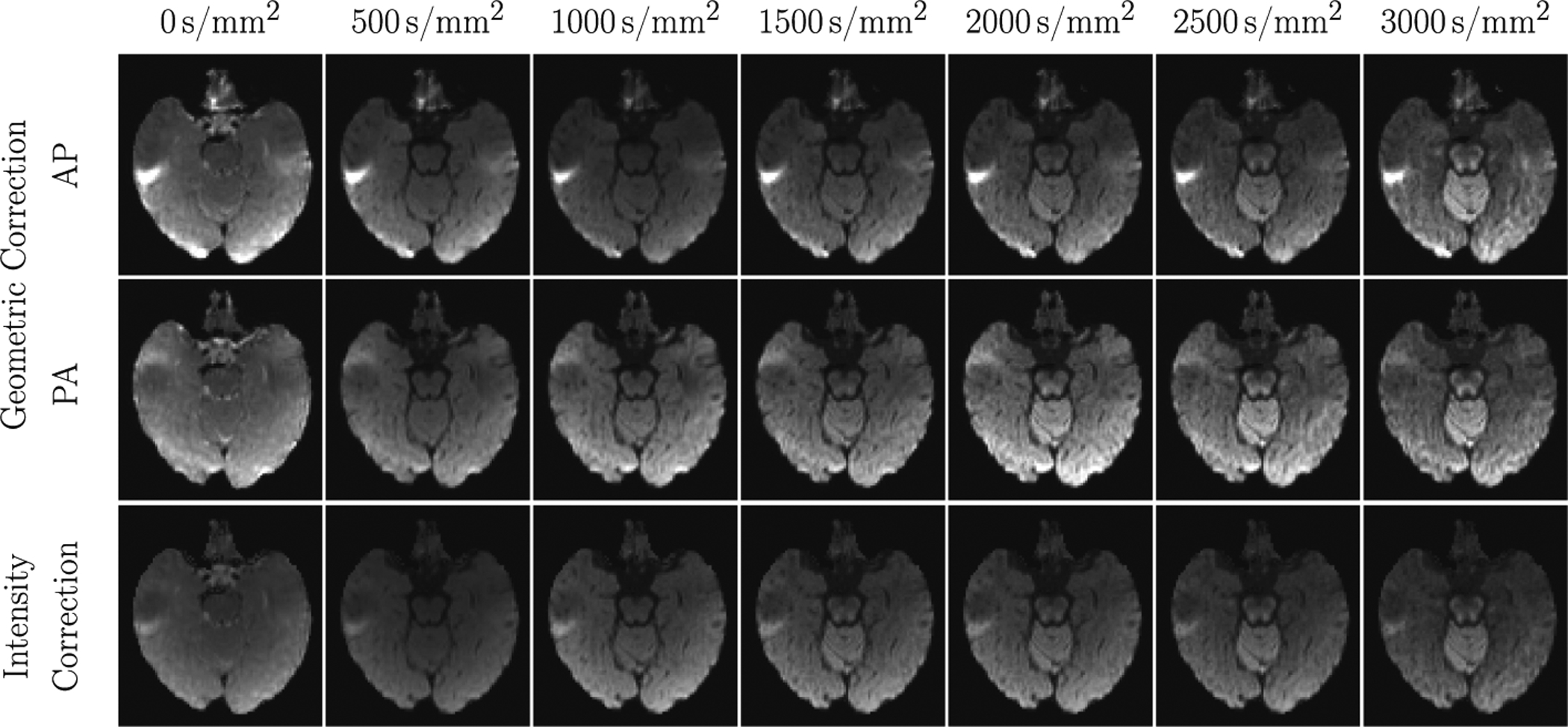
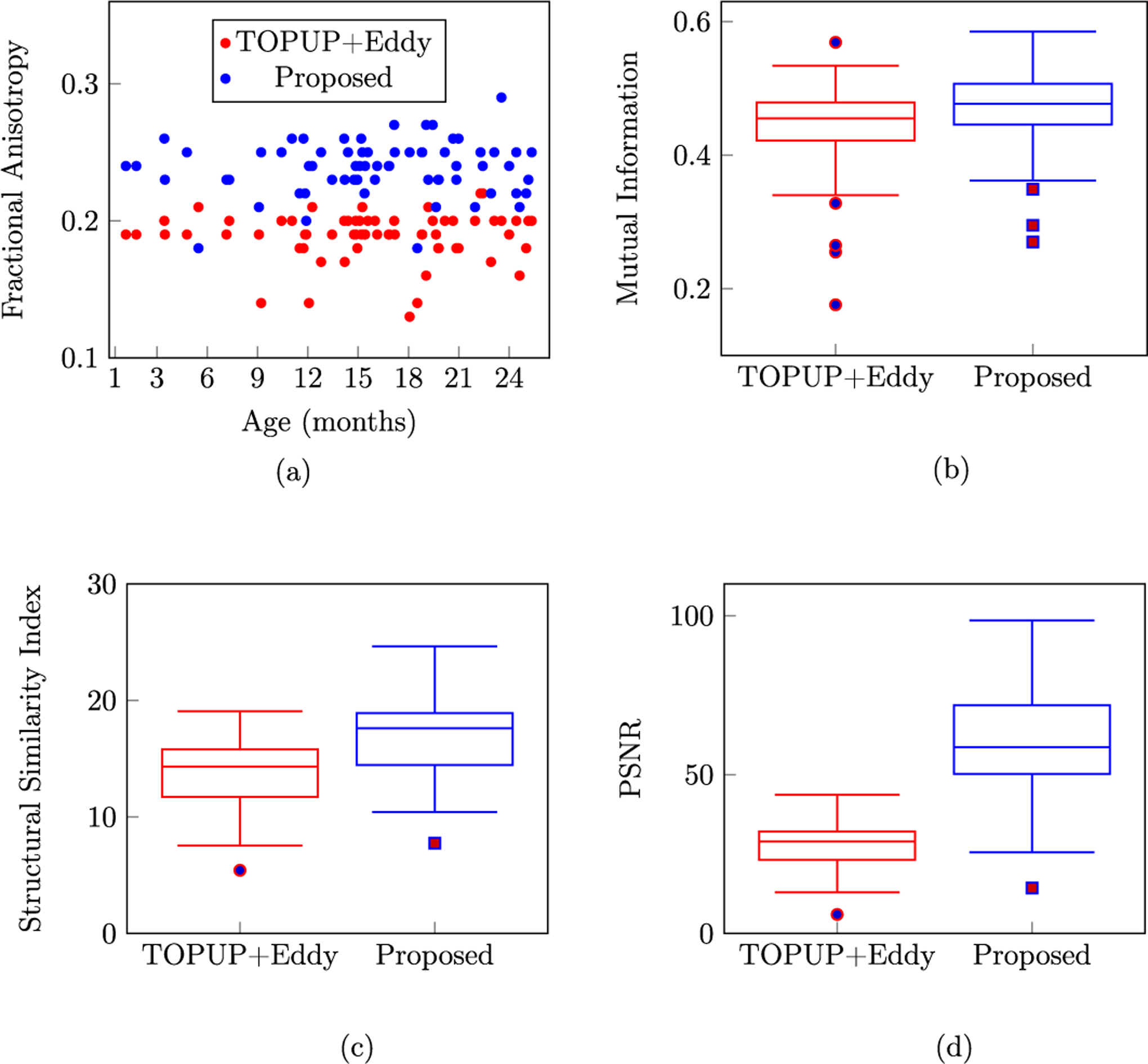
Ahmad, S., Wu, Y., Huynh, K. M., Thung, K. H., Lin, W., Shen, D., ... & UNC/UMN Baby Connectome Project Consortium. (2020,). Fast Correction of Eddy-Current and Susceptibility-Induced Distortions Using Rotation-Invariant Contrasts. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 34-43). Springer, Cham.